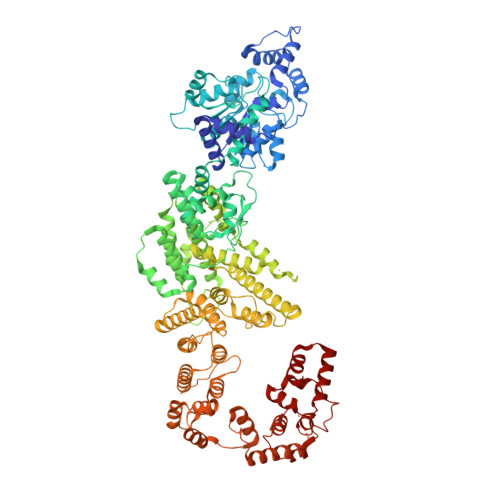
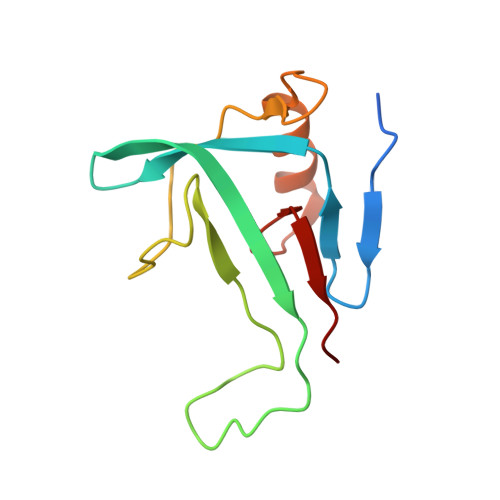
Insights into the modulation of bacterial NADase activity by phage proteins.
Yin, H., Li, X., Wang, X., Zhang, C., Gao, J., Yu, G., He, Q., Yang, J., Liu, X., Wei, Y., Li, Z., Zhang, H.(2024) Nat Commun 15: 2692-2692
- PubMed: 38538592
- DOI: https://doi.org/10.1038/s41467-024-47030-z
- Primary Citation of Related Structures:
8K98, 8K9A, 8W56, 8WKN, 8XKN - PubMed Abstract:
The Silent Information Regulator 2 (SIR2) protein is widely implicated in antiviral response by depleting the cellular metabolite NAD + . The defense-associated sirtuin 2 (DSR2) effector, a SIR2 domain-containing protein, protects bacteria from phage infection by depleting NAD + , while an anti-DSR2 protein (DSR anti-defense 1, DSAD1) is employed by some phages to evade this host defense. The NADase activity of DSR2 is unleashed by recognizing the phage tail tube protein (TTP). However, the activation and inhibition mechanisms of DSR2 are unclear. Here, we determine the cryo-EM structures of DSR2 in multiple states. DSR2 is arranged as a dimer of dimers, which is facilitated by the tetramerization of SIR2 domains. Moreover, the DSR2 assembly is essential for activating the NADase function. The activator TTP binding would trigger the opening of the catalytic pocket and the decoupling of the N-terminal SIR2 domain from the C-terminal domain (CTD) of DSR2. Importantly, we further show that the activation mechanism is conserved among other SIR2-dependent anti-phage systems. Interestingly, the inhibitor DSAD1 mimics TTP to trap DSR2, thus occupying the TTP-binding pocket and inhibiting the NADase function. Together, our results provide molecular insights into the regulatory mechanism of SIR2-dependent NAD + depletion in antiviral immunity.
Organizational Affiliation:
State Key Laboratory of Experimental Hematology, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, International Joint Laboratory of Ocular Diseases (Ministry of Education), Tianjin Key Laboratory of Ocular Trauma, School of Basic Medical Sciences, Tianjin Medical University, Tianjin, China.